Mathematical Oncology
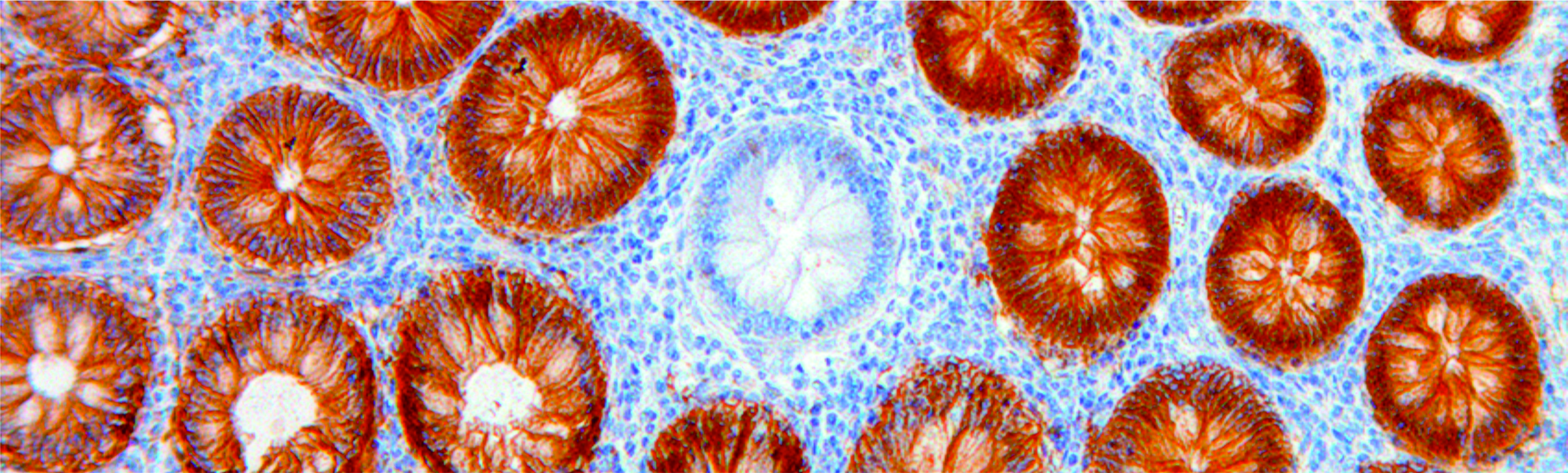
Cancer is one of the leading causes of disease-related death worldwide. In recent years, rapid increase in the molecular understanding of cancer has unraveled significant additional complexity of the disease. Although large amounts of data on cancer genetics and molecular characteristics are available and accumulating with increasing speed, adequate interpretation of these data still represents a major bottleneck. This is exactly where mathematics can be applied to oncology.
Through mathematical modeling of complex biological processes we are able to gain novel, unprecedented medical insights. The fields of application of mathematical models include the analysis of biological concepts and medical hypotheses about cancer evolution, and the prediction of clinical outcomes using existing clinical and molecular information. On the other hand, the medical applications give rise to mathematical challenges, which can lead to new methods and algorithms in various fields of mathematics, like data analysis, mathematical modeling and machine learning. Therefore, applying mathematics in the field of oncology will facilitate data interpretation and improve our understanding of carcinogenic processes.
Current projects
Publications / Output
Colorectal cancer incidences in Lynch syndrome: a comparison of results from the prospective lynch syndrome database and the international mismatch repair consortium. Hered Cancer Clin Pract. 2022 Oct 1;20(1):36. PMID: 36182917
S. Haupt, N. Gleim, A. Ahadova, H. Bläker, M. Knebel Doeberitz, M. Kloor, V. Heuveline: A computational model for investigating the evolution of colonic crypts during Lynch syndrome carcinogenesis. Computational and Systems Oncology, June 2021.
Mathematical modeling of multiple pathways in colorectal carcinogenesis using dynamical systems with Kronecker structure. PLoS Comput Biol. 2021 May 18;17(5):e1008970. PMID: 34003820
The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat Commun. 2020 Sep 21;11(1):4740. PMID: 32958755
. Age-dependent performance of BRAF mutation testing in Lynch syndrome diagnostics. Int J Cancer. 2020 Nov 15;147(10):2801-2810. PMID: 32875553
von Knebel Doeberitz M, Kloor M. Towards a vaccine to prevent cancer in Lynch syndrome patients.Fam Cancer. 2013 Jun;12(2):307-12. doi: 10.1007/s10689-013-9662-7. Review. PMID: 23760517
Reuschenbach M, Kloor M, Morak M, Wentzensen N, Germann A, Garbe Y, Tariverdian M, Findeisen P, Neumaier M, Holinski-Feder E, von Knebel Doeberitz M. Serum antibodies against frameshift peptides in microsatellite unstable colorectal cancer patients with Lynch syndrome.Fam Cancer. 2010 Jun;9(2):173-9. PMID: 19957108
Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, Tariverdian M, Benner A, von Knebel Doeberitz M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. 2008 Apr;134(4):988-97. PMID: 18395080